circRNA,IncRNA and mRNA in human blood exosomes
Introduction
exoRBase is a repository of circular RNA (circRNA), long non-coding RNA (lncRNA) and messenger RNA (mRNA) derived from RNA-seq data analyses of human blood exosomes. Experimental validations from published literature are also included.
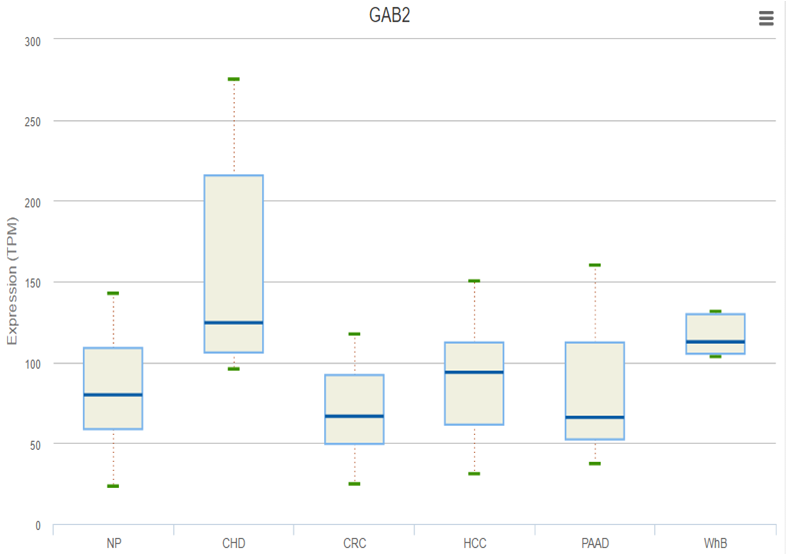
exoRBase features the integration and visualization of RNA expression profiles based on normalized RNA-seq data spanning both normal individuals and patients with different diseases.
exoRBase aims to collect and characterize all long RNA species in human blood exosomes. The annotation, expression level and possible original tissues are provided. exoRBase will aid researchers in identifying molecular signatures in blood exosomes and will trigger new circulating biomarker discovery and functional implication for human diseases.
Release Notes&News
- EV-origin: enumerating the tissue-cellular origin of circulating extracellular vesicles using exLR profile, published in Comput Struct Biotechnol J. [October 2020] [ Full Text]
- The exLR biomarkers used for the detection of pancreatic ductal adenocarcinoma published in Gut. [March 2020] [ Full Text]
- The feature and application of human blood exLRs published in Clin Chem. [March 2019] [Full Text]
- exoRBase has received 10,000 visitors. [May, 16 2018]
- exoRBase is published in Database Issue of Nucleic Acids Research. [October 2017] [Full Text]
- exoRBase is publicly accessible. [August 2017]
- 77 experimental validations from 18 published literatures have been included. [July 2017]
- GTEx and circBase data have been used to annotate possible original cells/tissues of exosomal RNAs. [July 2017]
- 92 RNA-seq samples from seven experiments have been integrated into exoRBase. [June 2017]