
circRNA,IncRNA and mRNA in human blood exosomes
Frequently Asked Questions
1. What is exoRbase?
exoRBase is a repository of circular RNA (circRNA), long non-coding RNA (lncRNA) and messenger RNA (mRNA) derived from RNA-seq data analysis on human blood exosomes. exoRBase features integration and visualization of RNA expression profiles based on normalized RNA-seq data spanning normal individuals and patients with different diseases.
2. How were collected RNA-seq datasets were processed?
All collected datasets were processed through a consistent bioinformatics pipeline. The raw sequencing reads were first trimmed by removing adapters and low-quality bases using Trimmomatic. Then the trimmed reads were mapped to human reference genome (GRCh38) by HISAT2. featureCounts were employed to quantify the number of reads aligned to regions of protein-coding (mRNAs) or long non-coding RNAs (lncRNAs). Read counts for each gene were normalized to TPM values (Transcript Per Million) . Annotation of protein-coding and long non-coding RNAs were retrieved from GENCODE v25. Additionally, circular RNAs were identified and quantified by ACFS and find_circ. Circular RNAs that were detected by both tools were considered in our analysis. Taken consideration of various library sizes, the read counts of each circular RNAs were normalized by calculating the RPM values (Read counts per Million mapped reads). Following is the flowchart of data process and overall design of exoRBase.
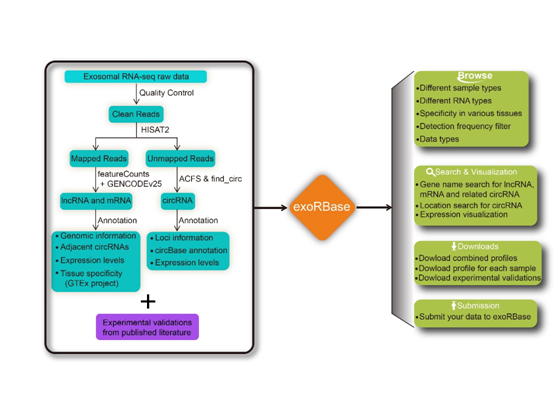
3. How were tissue specificity scores for lncRNA and mRNA calculated?
To assess tissue specificity of protein-coding and long non-coding RNAs across human tissues, we downloaded expression atlas from GTEx (Genotype-Tissue Expression project), which profiled gene expression across 30 human tissues. Genes that were lowly expressed (<0.1 RPKM) in all tissues were removed.We defined the tissue specificity score as the difference between the logarithm of the total number of tissues and the Shannon entropy of the expression values for a gene. The score was calculated as follows:
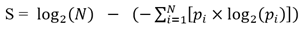
Where pi stands for the relative frequency, which was computed as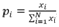 , Xi is the gene expression level for gene in tissue i, N is the total number of all tissues.
, Xi is the gene expression level for gene in tissue i, N is the total number of all tissues.
For each gene, one specificity score and 30 frequency scores (p_i) were calculated. A gene was defined as tissue-specific when its max frequency score was more than double the second largest one and its specificity score was no less than 1.
4. How were circular RNAs were assigned to their linear RNAs?
Circular RNAs are transcribed from intra- or inter-genic loci. A circular RNA was assigned to be related to a gene when this circular RNA was transcribed from regions within the gene or this gene was located nearest to the circular RNA.
5. What do the abbreviations of disease names stand for?
'NP' stands for normal person, 'CHD' stands for Coronary Heart Disease, 'CRC' stands for Colorectal Cancer, 'HCC' stands for Hepatocellular Carcinoma, 'PAAD' stands for Pancreatic Adenocarcinoma, 'BC' stands for Breast Cancer, 'WhB' stand for Whole Blood.
6. Were the expression values further processed in heatmaps?
Yes. The expression values were further processed before drawing heatmaps as following:
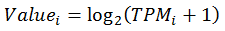
Where 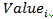 is the processed expression value used in heatmap for RNA i.
is the processed expression value used in heatmap for RNA i.
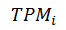 stands for the raw TPM value for RNA i.
stands for the raw TPM value for RNA i.
| Laboratory/Organization | Contributors | Data Samples | Data Status |
|---|---|---|---|
| Max Delbrück Center for Molecular Medicine, Berlin 13125, Germany | Sebastian Memczak,Nikolaus Rajewsky | Whole blood /exosomes | Published-public(GSE73570) |
| Institute of Biomedical Science, Fudan University, Shanghai 200032, China | Shengli Li, Shenglin Huang | Blood exosomes/ Normal person | Published-public(GSE100206) |
| Department of Biochemistry and Molecular Biology, Shanghai Medical College, Shanghai, 200032, China | Yan Tang | Blood exosomes /Coronary heart disease (CHD) | Unpublished-private (GSE99985) |
| Department of Colorectal Surgery, Fudan University Shanghai Cancer Center, Shanghai 200032, China | Jiang Zhao, Ye Xu | Blood exosomes /Colon cancer (CRC) | Unpublished-private (GSE100063) |
| Department of Hepatobiliary Surgery, Fudan University Shanghai Cancer Center, Shanghai 200032, China | Lu Wang | Blood exosomes /Hepatocellular carcinoma (HCC) | Unpublished-private (GSE100207) |
| Department of Integrative Oncology, Fudan University Shanghai Cancer Center, Shanghai 200032, China | Shulin Yu, Peng Wang | Blood exosomes /Pancreatic adenocarcinoma (PAAD) | Unpublished-private (GSE100232) |
| Department of Radiation Oncology, University of Pennsylvania, Philadelphia 19104, USA | Barzin Y. Nabet, Yu Qiu | Blood exosomes /Breast Cancer(BC) | Published-public(GSE93078) |
| Max Delbrück Center for Molecular Medicine, 13125 Berlin, Germany | Petar Glazar, Panagiotis Papavasileiou and Nikolaus Rajewsky | circRNA cell/tissue expression | Published-public(http://www.circbase.org/) |
| Institute of Biomedical Science, Fudan University, Shanghai 200032, China. | Qiupeng Zheng, Shenglin Huang | circRNA tissue expression | Published-public(GSE77661) |
| The Broad Institute of MIT and Harvard | GTEx prortal | lncRNA/mRNA tissue expression | Published-public (https://gtexportal.org/) |